Source code please see in Github repository
Q1 Fuzzy Clustering using EM
Given the training data EM Points.mat, you should implement the Fuzzy Clustering using EM algorithm for clustering.
The dataset contains 400 2D points totally with 2 clusters. Each point is in the format of [Xcoordinate, Y-coordinate, label].
You are required to implement Fuzzy Clustering using the EM algorithm.
- You are NOT allowed to use any existing EM library. You need to implement it manually and submit your code.
- Report the updated centers and SSE for the first two iterations. (If you set any hyper parameter when computing SSE, please write it clearly in the report.)
- Report the final converged centers for each cluster.
- In your report, draw the clustering results of your implemented algorithm and compare it with the original labels in the dataset. You need to discuss the result briefly.
Hint: For terminate condition, you can consider the change of parameters or the max iterations.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from scipy.io import loadmatpoints = loadmat("EM_Points.mat")["Points"]
pointsarray([[-0.3005007 , 0.71622977, 0. ],
[-0.07260801, 1.39849283, 0. ],
[-0.209947 , 1.32075583, 0. ],
...,
[ 1.42954638, 0.15714956, 1. ],
[ 1.14187298, -0.21691986, 1. ],
[ 1.49161487, -0.50410824, 1. ]])
Assume $o_i$ represents the i-th object and $c_j$ represents the j-th center. The sum of the squared error is computed by
$$ SSE(C) = \sum_{i=1}^{n} \sum_{j=1}^{k} w_{ij}^2 \cdot dist(o_i, c_j)^2 $$
class FuzzyClusteringEM:
def __init__(self, n_clusters, init_centroid, iteration):
self.n_clusters = n_clusters
self.init_centroid = init_centroid
self.iteration = iteration
def fit(self, X):
c0 = self.init_centroid[0]
c1 = self.init_centroid[1]
M = np.zeros((2, X.shape[0]))
for i in range(self.iteration):
# calculate the distance from object to center
dist_c0 = np.sum(np.square(X[:,:2] - c0), axis=1) # (n,)
dist_c1 = np.sum(np.square(X[:,:2] - c1), axis=1) # (n,)
dist = dist_c0 + dist_c1
# calculate M matrix
M = np.vstack((dist_c1/dist, dist_c0/dist)) # 2*n
# calculate the new centroids
c = np.square(M).dot(X[:,:2]) / np.sum(np.square(M), axis=1, keepdims=True)
# compute SSE (sum of squared error)
SSE = np.sum(np.square(M) * np.vstack((dist_c0, dist_c1)))
# print the results
print("iteration = %s" % i)
print("SSE = %s" % SSE)
print("C = ")
print(c)
print()
# update new centroids
c0 = c[0]
c1 = c[1]
# Store the predicted labels
self.labels_ = np.argmax(M, axis=0)
initc = np.array([[1,1],[2,2]])
clustering = FuzzyClusteringEM(n_clusters=2, init_centroid=initc, iteration=15)
clustering.fit(points)iteration = 0
SSE = 452.2064066190702
C =
[[0.53866521 0.55193537]
[0.47385793 0.60978921]]
iteration = 1
SSE = 209.93907719136445
C =
[[0.60141888 0.46557221]
[0.43167368 0.63543004]]
iteration = 2
SSE = 205.38336884149535
C =
[[0.7256404 0.32912566]
[0.3051053 0.77335614]]
iteration = 3
SSE = 183.55466797593803
C =
[[0.90598324 0.1294149 ]
[0.11855499 0.9793926 ]]
iteration = 4
SSE = 148.13131455149812
C =
[[1.01083941 0.01034638]
[0.00611246 1.09897161]]
iteration = 5
SSE = 137.8432366754871
C =
[[ 1.03788056 -0.02172753]
[-0.02550994 1.12770828]]
iteration = 6
SSE = 137.1701001910947
C =
[[ 1.04306219 -0.02775795]
[-0.03212375 1.13224185]]
iteration = 7
SSE = 137.14633312924246
C =
[[ 1.0440722 -0.02874075]
[-0.03347604 1.13280377]]
iteration = 8
SSE = 137.14555718757154
C =
[[ 1.04429125 -0.02887354]
[-0.03377623 1.13283733]]
iteration = 9
SSE = 137.14552695547323
C =
[[ 1.04434439 -0.02888064]
[-0.03385258 1.13282462]]
iteration = 10
SSE = 137.14552517019524
C =
[[ 1.04435838 -0.02887534]
[-0.03387519 1.13281695]]
iteration = 11
SSE = 137.14552500427823
C =
[[ 1.04436222 -0.02887173]
[-0.0338828 1.13281404]]
iteration = 12
SSE = 137.14552498415927
C =
[[ 1.04436327 -0.02886999]
[-0.0338856 1.13281311]]
iteration = 13
SSE = 137.1455249813707
C =
[[ 1.04436355 -0.02886921]
[-0.03388668 1.13281284]]
iteration = 14
SSE = 137.1455249809489
C =
[[ 1.04436361 -0.02886888]
[-0.03388712 1.13281277]]
We set two initial centroids
$$ C_1 = (1,1), C_2 = (2,2) $$
In the first three iterations, the updated centers and SSE
| Iteration | Updated Centers | SSE |
|---|---|---|
| 0 | $$C_1=(0.539,0.552),C_2=(0.474,0.610)$$ | 452.206 |
| 1 | $$C_1=(0.601,0.466),C_2=(0.432,0.635)$$ | 209.939 |
| 2 | $$C_1=(0.726,0.329),C_2=(0.305,0.773)$$ | 205.383 |
After about 10 iterations, we can get the final converged centers
$$ C_1 = (1.04,-0.0289), C_2 = (-0.0339,1.133) $$
with $SSE = 137.145$
pred_labels = clustering.labels_
points_pred = np.hstack((points[:,:2], pred_labels.reshape(-1,1))) # hstack points with predicted labels
dfA = pd.DataFrame(points_pred) # points with predicted labels
dfB = pd.DataFrame(points) # points with original labelsfig = plt.figure(figsize=(15,4))
fig1 = plt.subplot(1,2,1)
fig2 = plt.subplot(1,2,2)
# plot sub figure 1
plt.sca(fig1)
plt.title("Predicted Clustering by Fuzzy Clutering with EM")
plt.xlabel('x')
plt.ylabel('y')
area = np.pi * 4**2
x0 = dfA[dfA[2]==0][0]
y0 = dfA[dfA[2]==0][1]
x1 = dfA[dfA[2]==1][0]
y1 = dfA[dfA[2]==1][1]
plt.scatter(x0, y0, s=area, c='#00CED1', alpha=0.4, label='Cluster A')
plt.scatter(x1, y1, s=area, c='#DC143C', alpha=0.4, label='Cluster B')
plt.legend()
# plot sub figure 2
plt.sca(fig2)
plt.title("Original Clutering")
plt.xlabel('x')
plt.ylabel('y')
area = np.pi * 4**2
x0 = dfB[dfB[2]==0][0]
y0 = dfB[dfB[2]==0][1]
x1 = dfB[dfB[2]==1][0]
y1 = dfB[dfB[2]==1][1]
plt.scatter(x0, y0, s=area, c='#DC143C', alpha=0.4, label='Cluster A')
plt.scatter(x1, y1, s=area, c='#00CED1', alpha=0.4, label='Cluster B')
plt.legend()
plt.show()According to the figures we draw, we can clearly see that predicted clustering has a more distinct boundary than the clutering divided by original labels. I have drawed the clutering boundary below:
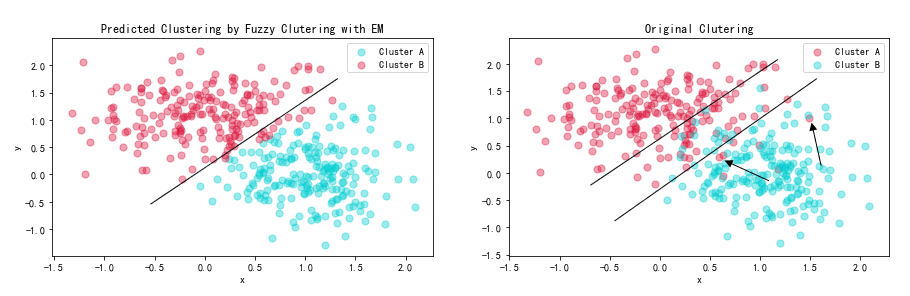
We can easily draw a line to distinguish two cluters in the left figure. However, in the figure on the right, though we widen the boundary, there are still some outliers existing. This is because in reality, the noise exists which would make some points fluctuate near the classification boundary. However, The algorithm just consider the ideal situation and cannot consider noise.
Q2 DBSCAN
Given the dataset DBSCAN.mat with 500 2D points, you should apply DBSCAN algorithm to
cluster the dataset and find outliers as the following settings:
Parameter Setting
- Set $\epsilon$ = 5, Minpoints=5.
- Set $\epsilon$ = 5, Minpoints=10
- Set $\epsilon$ = 10, Minpoints=5.
- Set $\epsilon$ = 10, Minpoints=10.
Implementation
- Draw a picture for your cluster results and outliers in each parameter setting in your report. For clearly, in each picture, the color of outliers should be BLUE.
- Add a table to report how many clusters and outliers you find in each parameter setting in your report.
- Discuss the results of different parameter settings, and report the best setting that you think and write your reason clearly.
- Note that you are NOT allowed to use any existing DBSCAN library. You need to submit your code.
import random
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from scipy.io import loadmatpoints = loadmat("DBSCAN.mat")["Points"]class DBSCAN:
def __init__(self, eps, min_points):
self.eps = eps
self.min_points = min_points
def find_neigh(self, j, X):
"""
Find all eps-neighborhood of point j
"""
N = []
for p in range(X.shape[0]):
# compute the Euclidean distance
temp = np.sqrt(np.sum(np.square(X[j] - X[p])))
if(temp <= self.eps):
N.append(p)
return N
def fit(self, X):
"""
Perform DBSCAN on data X
It will save the labels, n_cluters and n_outliers as attributes
"""
k = -1
n_outliers = 0
neigh_pts = [] # neighbor points
ner_neighb_pts = [] # near neighbor points
visited = [] # set of points that have been visited
no_visit = [x for x in range(len(X))] # list of points that not have been visited
cluster = [-1 for y in range(len(X))] # initialize clusters, all points are set to be outliers
while len(no_visit) > 0:
j = random.choice(no_visit)
no_visit.remove(j) # delete j point from no_visit list
visited.append(j) # add the j point in visited set
neigh_pts = self.find_neigh(j, X)
if len(neigh_pts) < self.min_points:
cluster[j] = -1 # the point is outlier (also represent color blue in the sequnce of colormap)
n_outliers += 1
else:
k += 1
cluster[j] = k
for i in neigh_pts:
if i not in set(visited):
no_visit.remove(i) # remove the point from no_visit list
visited.append(i) # add the point to visited list
ner_neigh_pts = self.find_neigh(i, X) # find the near neighbors
if len(ner_neigh_pts) >= self.min_points:
for a in ner_neigh_pts:
if a not in neigh_pts:
neigh_pts.append(a)
if (cluster[i] == -1):
cluster[i] = k
self.labels_ = cluster
self.n_clusters_ = k + 1
self.n_outliers_ = n_outlierseps_list = [5, 10]
min_points_list = [5, 10]
for eps in eps_list:
for min_points in min_points_list:
# DBSCAN clustering
clustering = DBSCAN(eps, min_points)
clustering.fit(points)
# print the number of clusters and outliers
print("n_cluters = %s" % clustering.n_clusters_)
print("n_outliers = %s" % clustering.n_outliers_)
# plot the clutering scatter
area = np.pi * 4**2
plt.title("eps = %s and min_points = %s" %(eps, min_points))
plt.xlabel('x')
plt.ylabel('y')
plt.scatter(points[:,0], points[:,1], c=clustering.labels_, alpha=0.9)
plt.show()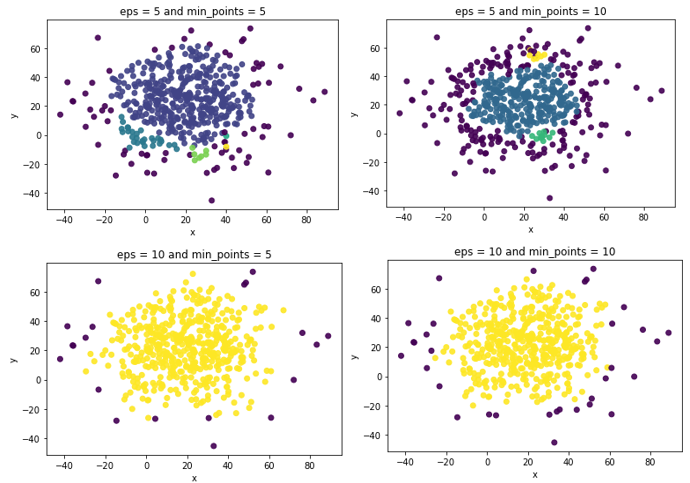
From the figures above, we could see that the outliers are mainly distributed in the edge area, which shown in dark BLUE. When using parameters eps = 10 and min_points = 5, the clusters are clear and distinct with less number of outliers. If the parameters are set as eps = 5 and min_points = 10, the number of outliers are up to 198. Not very well.
The detailed results are shown in the table:
| eps | min_points | n_clusters | n_outliers |
|---|---|---|---|
| 5 | 5 | 5 | 71 |
| 5 | 10 | 3 | 198 |
| 10 | 5 | 1 | 20 |
| 10 | 10 | 1 | 33 |
本博客文章除特别声明外,均可自由转载与引用,转载请标注原文出处:http://www.yelbee.top/index.php/archives/168/
